Jan 11, 2021
rOpenSci HQ
Our own Maëlle Salmon was featured in an US-RSE Stories podcast. The US-RSE is The United States (US) Research Software Engineer Association. This is a great chance to learn more about Maëlle and some of the work she does.
Community Calls
Our next Community Call will cover Statistical Software Peer Review. The date is not yet set. See the link for more details.
Code of Conduct
Make sure to read our two recent posts on our Code of Conduct: rOpenSci Code of Conduct Annual Review and rOpenSci 2020 Code of Conduct Transparency Report
Software 📦
CRAN: ![]() GitHub:
GitHub: ![]()
New Versions
- A new version (
v1.2.1) ofcld2is on CRAN - R wrapper for Google’s Compact Language Detector 2. See the release notes for changes. Checkout the docs to get started.

- A new version (
v1.4.1) ofcld3is on CRAN - Bindings to Google’s Compact Language Detector 3. See the release notes for changes. Checkout the docs to get started.

- A new version (
v0.28.0) ofgit2ris on CRAN - an interface to the libgit2 library, a pure C implementation of the Git core methods. See the release notes for changes. Checkout the docs to get started.

- A new version (
v4.9.3) ofplotlyis on CRAN - Create Interactive Web Graphics via plotly.js. See the NEWS for changes. Checkout the plotly book to get started.

- A new version (
v0.3.0) ofPostcodesioRis on CRAN - API wrapper for Postcodes.io. See the release notes for changes. Checkout the docs to get started.

- A new version (
v1.3.0) ofrnoaais on CRAN - NOAA Weather Data from R. See the release notes for changes. Checkout the docs to get started.

- A new version (
v0.8.1) ofstplanris on CRAN - sustainable transport planning. See the release notes for changes. Checkout the docs to get started.

- A new version (
v0.7.2) ofwellknownis on CRAN - Convert between WKT and GeoJSON. See the release notes for changes. Checkout the docs to get started.

- A new version (
v1.0.3) ofplateris on CRAN - read, tidy, and display data from microtiter plates. See the release notes for changes. Checkout the docs to get started.

- A new version (
v3.4.2) ofrgbifis on CRAN - interface to the Global Biodiversity Information Facility API. See the release notes for changes. Checkout the docs to get started.

- A new version (
v1.2.0) ofspoccis on CRAN - an interface to many species occurrence data sources. See the release notes for changes. Checkout the docs to get started.

- A new version (
v7.13.0) ofdrakeis on CRAN - a pipeline toolkit for reproducible computation at scale. See the release notes for changes. Checkout the docs to get started.

- A new version (
v1.1-6) ofsuppdatais on CRAN - downloading supplementary data from published manuscripts. See the release notes for changes. Checkout the docs to get started.

Software Review ✔
We accept community contributed packages via our software review system - an open software review system, sorta like scholarly paper review, but way better. We’ll highlight newly onboarded packages here. A huge thanks to our reviewers, who do a lot of work reviewing (see the blog post on our review system), and the authors of the packages!
If you want to be a reviewer fill out this short form, and we’ll ping you when there’s a submission that fits in your area of expertise.
The following package was recently submitted:
- epair > Grabs data from EPA API, simplifies getting pollutant data
- Author: G.L. Orozco-Mulfinger
- Issue: ropensci/software-review#418
- Reviewers: not yet assigned
On the blog
- Thank You to the rOpenSci Community, 2020 by Stefanie Butland and Steffi LaZerte
- Donate to rOpenSci this Giving Season by Karthik Ram
- 2 Months in 2 Minutes - rOpenSci News, December 2020 by Stefanie Butland
- rOpenSci Code of Conduct Annual Review by Stefanie Butland, Scott Chamberlain, Mark Padgham, and Kara Woo
- rOpenSci 2020 Code of Conduct Transparency Report by Stefanie Butland, Scott Chamberlain, and Kara Woo
Software review
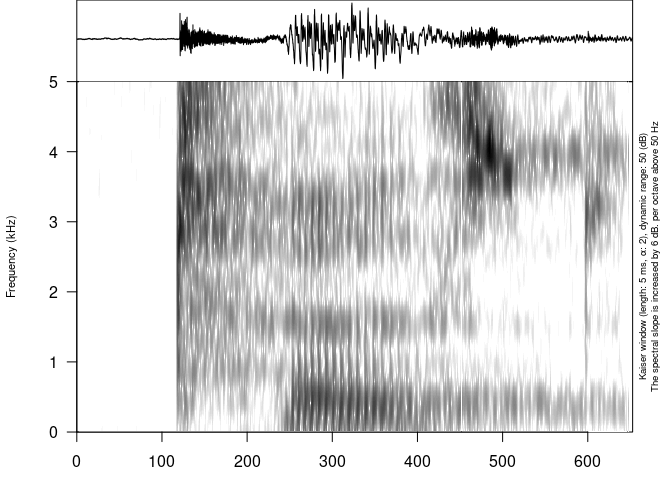
- George Moroz wrote about his package phonfieldwork that recently went through rOpenSci software review: Phonetic Fieldwork and Experiments with the phonfieldwork Package for R
Tech notes
- Scott Chamberlain wrote a post on HTTP Testing With the Newest Release of vcr, covering a new version of vcr.
Citations
The following 25 works use/cite rOpenSci software:
- Grattarola & Rodríguez-Tricot used taxize in their paper Mammals of Paso Centurión, an area with relicts of Atlantic Forest in Uruguay 1
- Calderón Franco used taxize in their paper Hi-C pipeline development for linking resistome and mobilome to the microbiome of wastewater samples 2
- Hansen & Jansa used textreuse in their paper Complexity, Resources, and Text Borrowing in State Legislatures 3
- Ilík used taxize in their paper High-throughput sequencing as a tool for studying strongylid nematode communities in primates 4
- Walton et al. used taxize in their paper Landscape Analysis for the Specimen Data Refinery 5
- Ossola et al. used taxize in their paper The Global Urban Tree Inventory: A database of the diverse tree flora that inhabits the world’s cities 6
- Atwood et al. used taxize in their paper Herbivores at the highest risk of extinction among mammals, birds, and reptiles 7
- Morgan & Lovelace used stplanr in their paper Travel flow aggregation: Nationally scalable methods for interactive and online visualisation of transport behaviour at the road network level 8
- Carroll et al. used treeio in their paper Twentieth-century emergence of antimicrobial resistant human- and bovine-associated Salmonella enterica serotype Typhimurium lineages in New York State 9
- Whitmer et al. used treeio in their paper c(“Inference of Nipah virus Evolution”, “Virus Evolution”) 10
- Ettinger & Eisen used treeio in their paper Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina 11
- Grassl et al. used tokenizers in their paper Dark and bright patterns in cookie consent requests 12
- Scott et al. used tokenizers in their paper NEPA and National Trends in Federal Infrastructure Siting in the United States 13
- López Galán et al. used tokenizers in their paper Dynamic Courtship Signals and Mate Preferences in Sepia plangon 14
- Brandão et al. used tokenizers in their paper PlatCOVID: A Novel Web Tool to Analyze, Curate and Share COVID-19 Literature 15
- Wang et al. used UCSCXenaTools in their paper UCSCXenaShiny: An R Package for Exploring and Analyzing UCSC Xena Public Datasets in Web Browser 16
- Obradovich et al. used rnaturalearth in their paper Expanding the Measurement of Culture with a Sample of Two Billion Humans 17
- Gutiérrez-Hernández used MODIStsp in their paper Fenología de los ecosistemas de alta montaña en Andalucía: Análisis de la tendencia estacional del SAVI (2000-2019 18
- Hazlett et al. used refsplitr in their paper The geography of publishing in the Anthropocene 19
- Mothes et al. used rredlist and natserv in their paper Protect or perish: Quantitative analysis of state‐level species protection supports preservation of the Endangered Species Act 20
- Falconi et al. used rgbif in their paper An open-access occurrence database for Andean bears in Peru 21
- Costanzi et al. used webchem in their paper Lists of Chemical Warfare Agents and Precursors from International Nonproliferation Frameworks: Structural Annotation and Chemical Fingerprint Analysis 22
- Batista et al. used rentrez in their paper Botryosphaeriaceae species on forest trees in Portugal: diversity, distribution and pathogenicity 23
- Rasher et al. used rnoaa in their paper Keystone predators govern the pathway and pace of climate impacts in a subarctic marine ecosystem 24
- Kuchta et al. used rotl in their paper Diversity of monogeneans and tapeworms in cypriniform fishes across two continents 25
From the Forum
We have a discussion forum (using Discourse) for the rOpenSci community. It’s a really nice way to have conversations on the internet. From time to time we’ll highlight recent discussions of interest.
Two new use cases were shared in the forum over the past month since our last newsletter:
- Isabella Velásquez shared: Scraping liked posts on Twitter using rtweet; the use case employed the rtweet package, an rOpenSci package maintained by Michael Kearney
- Maëlle Salmon shared: Using av to convert audio files for compatibility with an electronic storyteller; the use case employed the av package, an rOpenSci package maintained by Jeroen Ooms
Call For Maintainers
Part of the mission of rOpenSci is making sustainable software that users can rely on. Some software maintainers need to give up maintenance due to a variety of circumstances. When that happens we try to find new maintainers. Check out our guidance for taking over maintenance of a package.
We’ve had eight recent examples of maintainer transitions within rOpenSci:
- RSelenium: now maintained by Ju Kim
- chromer: now maintained by Paula Andrea
- qualtRics: now maintained by Julia Silge
- rsnps: now maintained by Julia Gustavsen and Sina Rüeger
- webchem: now maintained by Erik Sapper
- mregions: now maintained by Lennert Schepers (VLIZ)
- rWBclimate: now maintained by Sergio Ibarra-Espinosa and Amanda Rehbein
- rinat: now maintained by Stéphane Guillou
We’ve got three packages in need of a new maintainer. The current maintainer of all three packages is looking for new maintainers. Email Scott if you’re interested. If a new maintainer is not found these packages will be archived.
- traits: species trait data from around the web
- getlandsat: download Landsat 8 data
- originr: Fetch species origin data from the web
Get involved with rOpenSci
We maintain a Contributing Guide that can help direct you to the right place, whether you want to make code contributions, non-code contributions, or other things like sharing use cases.
Keep up with rOpenSci
- Mailing list: Sign up with an email address to get this newsletter sent to your inbox -> https://news.ropensci.org/
- Alternatively, you can subscribe to this newsletter via our XML feed at https://news.ropensci.org/feed.xml or our JSON feed at https://news.ropensci.org/feed.json
- rOpenSci on Twitter: @ropensci
- The rOpenSci blog at ropensci.org/blog - you can subscribe in any RSS aggregator, or manually via https://ropensci.org/index.xml. We also announce new blog posts on our Twitter account.
Footnotes
-
Grattarola, F., & Rodríguez-Tricot, L. (2020). Mammals of Paso Centurión, an area with relicts of Atlantic Forest in Uruguay. Neotropical Biology and Conservation, 15(3), 267–283. https://doi.org/10.3897/neotropical.15.e53062 ↩
-
Calderón Franco, D. (2020). Hi-C pipeline development for linking resistome and mobilome to the microbiome of wastewater samples https://bit.ly/3i7Q59d ↩
-
Hansen, E. R., & Jansa, J. M. (2020). Complexity, Resources, and Text Borrowing in State Legislatures. https://bit.ly/3nvLK0R ↩
-
Ilík, B. V. High-throughput sequencing as a tool for studying strongylid nematode communities in primates. Thesis. https://is.muni.cz/th/vg2vb/DP_Ilik_Vladislav.pdf ↩
-
Walton, S., Livermore, L., Bánki, O., Cubey, R., Drinkwater, R., Englund, M., … Wu, Z. (2020). Landscape Analysis for the Specimen Data Refinery. Research Ideas and Outcomes, 6. https://doi.org/10.3897/rio.6.e57602 ↩
-
Ossola, A., Hoeppner, M. J., Burley, H. M., Gallagher, R. V., Beaumont, L. J., & Leishman, M. R. (2020). The Global Urban Tree Inventory: A database of the diverse tree flora that inhabits the world’s cities. Global Ecology and Biogeography, 29(11), 1907–1914. https://doi.org/10.1111/geb.13169 ↩
-
Atwood, T. B., Valentine, S. A., Hammill, E., McCauley, D. J., Madin, E. M. P., Beard, K. H., & Pearse, W. D. (2020). Herbivores at the highest risk of extinction among mammals, birds, and reptiles. Science Advances, 6(32), eabb8458. https://doi.org/10.1126/sciadv.abb8458 ↩
-
Morgan, M., & Lovelace, R. (2020). Travel flow aggregation: Nationally scalable methods for interactive and online visualisation of transport behaviour at the road network level. Environment and Planning B: Urban Analytics and City Science, 239980832094277. https://doi.org/10.1177/2399808320942779 ↩
-
Carroll, L. M., Huisman, J. S., & Wiedmann, M. (2020). Twentieth-century emergence of antimicrobial resistant human- and bovine-associated Salmonella enterica serotype Typhimurium lineages in New York State. Scientific Reports, 10(1). https://doi.org/10.1038/s41598-020-71344-9 ↩
-
Whitmer, S. L. M., Lo, M. K., Sazzad, H. M. S., Zufan, S., Gurley, E. S., Sultana, S., … Klena, J. D. (2020). Inference of Nipah virus Evolution, 1999-2015. Virus Evolution. https://doi.org/10.1093/ve/veaa062 ↩
-
Ettinger, C. L., & Eisen, J. A. (2020). Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PLOS ONE, 15(7), e0236135. https://doi.org/10.1371/journal.pone.0236135 ↩
-
Grassl, P., Schraffenberger, H., Zuiderveen Borgesius, F., & Buijzen, M. (2020, July 21). Dark and bright patterns in cookie consent requests. https://doi.org/10.31234/osf.io/gqs5h ↩
-
Scott, T. A., Ulibarri, N., & Perez Figueroa, O. (2020). NEPA and National Trends in Federal Infrastructure Siting in the United States. Review of Policy Research. https://doi.org/10.1111/ropr.12399 ↩
-
López Galán, A., Chung, W.-S., & Marshall, N. J. (2020). Dynamic Courtship Signals and Mate Preferences in Sepia plangon. Frontiers in Physiology, 11. https://doi.org/10.3389/fphys.2020.00845 ↩
-
Brandão, L. A. C., Agrelli, A., Bernardo, L., Paparella, F., Moura, R., & Crovella, S. (2020). PlatCOVID: A Novel Web Tool to Analyze, Curate and Share COVID-19 Literature. https://doi.org/10.21203/rs.3.rs-42169/v1 ↩
-
Wang, S., Xiong, Y., Gu, K., Zhao, L., Li, Y., Zhao, F., … Liu, X.-S. (2020). UCSCXenaShiny: An R Package for Exploring and Analyzing UCSC Xena Public Datasets in Web Browser. https://doi.org/10.20944/preprints202007.0179.v1 ↩
-
Obradovich, N., Özak, Ö., Martín, I., Ortuño-Ortín, I., Awad, E., Cebrián, M., … Cuevas, Á. (2020). Expanding the Measurement of Culture with a Sample of Two Billion Humans. National Bureau of Economic Research. https://doi.org/10.3386/w27827 ↩
-
Gutiérrez-Hernández, O. (2020). Fenología de los ecosistemas de alta montaña en Andalucía: Análisis de la tendencia estacional del SAVI (2000-2019). Pirineos, 175, 055. https://doi.org/10.3989/pirineos.2020.175005 ↩
-
Hazlett, M. A., Henderson, K. M., Zeitzer, I. F., & Drew, J. A. (2020). The geography of publishing in the Anthropocene. Conservation Science and Practice, 2(10). https://doi.org/10.1111/csp2.270 ↩
-
Mothes, C. C., Stemle, L. R., Fonseca, T. N., Clements, S. L., Howell, H. J., & Searcy, C. A. (2020). Protect or perish: Quantitative analysis of state‐level species protection supports preservation of the Endangered Species Act. Conservation Letters. https://doi.org/10.1111/conl.12761 ↩
-
Falconi, N., Fuller, T. K., DeStefano, S., & Organ, J. F. (2020). An open-access occurrence database for Andean bears in Peru. Ursus, 2020(31e11), 1. https://doi.org/10.2192/ursus-d-19-00012.1 ↩
-
Costanzi, S., Slavick, C. K., Hutcheson, B. O., Koblentz, G. D., & Cupitt, R. T. (2020). Lists of Chemical Warfare Agents and Precursors from International Nonproliferation Frameworks: Structural Annotation and Chemical Fingerprint Analysis. Journal of Chemical Information and Modeling, 60(10), 4804–4816. https://doi.org/10.1021/acs.jcim.0c00896 ↩
-
Batista, E., Lopes, A., & Alves, A. (2020). Botryosphaeriaceae species on forest trees in Portugal: diversity, distribution and pathogenicity. European Journal of Plant Pathology, 158(3), 693–720. https://doi.org/10.1007/s10658-020-02112-8 ↩
-
Rasher, D. B., Steneck, R. S., Halfar, J., Kroeker, K. J., Ries, J. B., Tinker, M. T., … Estes, J. A. (2020). Keystone predators govern the pathway and pace of climate impacts in a subarctic marine ecosystem. Science, 369(6509), 1351–1354. https://doi.org/10.1126/science.aav7515 ↩
-
Kuchta, R., Řehulková, E., Francová, K., Scholz, T., Morand, S., & Šimková, A. (2020). Diversity of monogeneans and tapeworms in cypriniform fishes across two continents. International Journal for Parasitology, 50(10-11), 771–786. https://doi.org/10.1016/j.ijpara.2020.06.005 ↩